The development of multi-cellular organisms involves changes of tissue structure, where groups of cells differentiate, change shape and move to form new tissues or organs. While many genes and molecules involved in these and other biological processes have been identified, the mechanical basis of each sequence of cell shape changes remains elusive.
The aim of our group is to understand and eventually connect these sequence of events. We are interested in how life organisms shape themselves: from the cytoskeletal arrangement and deployment of molecular motors inside the cells, to the resulting cell-cell interactions that determine tissue shape and function.
Models of cytoskeletal contraction mechanisms
One major focus of our group is trying to understand the many mechanisms by which cytoskeletal networks contract and generate forces.
These models are simulated with the CytoSim software (www.cytosim.org)
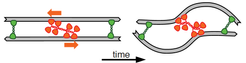
The best known mechanism of cytoskeletal contraction is the filament buckling, which relies on the combined actions of myosin motors and crosslinkers.
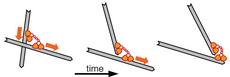
Cytoskeletal networks can also contract via other mechanism, such as the polarity sorting, which relies on the filament end-dwelling property of microtubule motors such as dynein and kinesin.
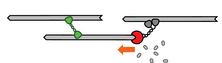
Another mechanism is the depolimerization end-tracking, where networks contract in the presence of a crosslinker that harness the the free energy of disassembling actin filaments.
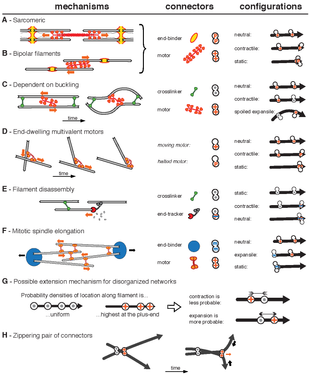
These 3 mechanism are only a subset of all the possible ways that cytoskeletal networks can contract. The image on the right show other theoretical mechanism of contraction and expansion of networks.
Our group is interested in exploring these different mechanism under different biological contexts and how they contribute to force generation and transmission within tissues, both at homeostasis and during development.
Models of tissue morphogenesis
Another area of activity in the group is the modelling of developmental processes at the cell and tissue scales. Our aim is to find out which set of cell behaviors and external constraints are necessary and sufficient to reproduce the wide variety of patterned structures in animal anatomy.
These models are simulated with the CompuCell3D software (compucell3d.org).
Somitogenesis and the Clock and Wavefront model
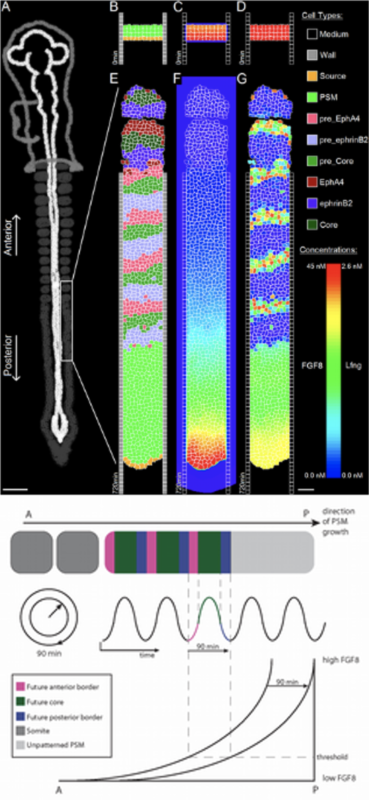
Many structures of our bodies, such as our vertebrae, ribs and skeletal muscles, are preceded from transient structures called somites. This process (somitogenesis) happens as our bodies elongate and each somite is created at regular temporal and spatial sequence from a growing and undifferentiated tissue called PSM (pre-somitic mesoderm).
The clock and wavefront model postulates that the interaction of a moving wavefront at the tissue level and an oscillating molecular clock, at the cellular level, is sufficient to determine the placement of the boundaries between the somites (figure on the left).
Somite segmentation as a self-organized process
In a notable work by Claudio Stern and colleagues, quail somites were induced to form simultaneously, without the presence of any internal molecular clock or external wavefront.
To further support their finding, we showed through simulations that cells undergoing mesenchymal-to-epitheial transition (polarization + elongation + apical constriction) can form somite-like structures autonomously.
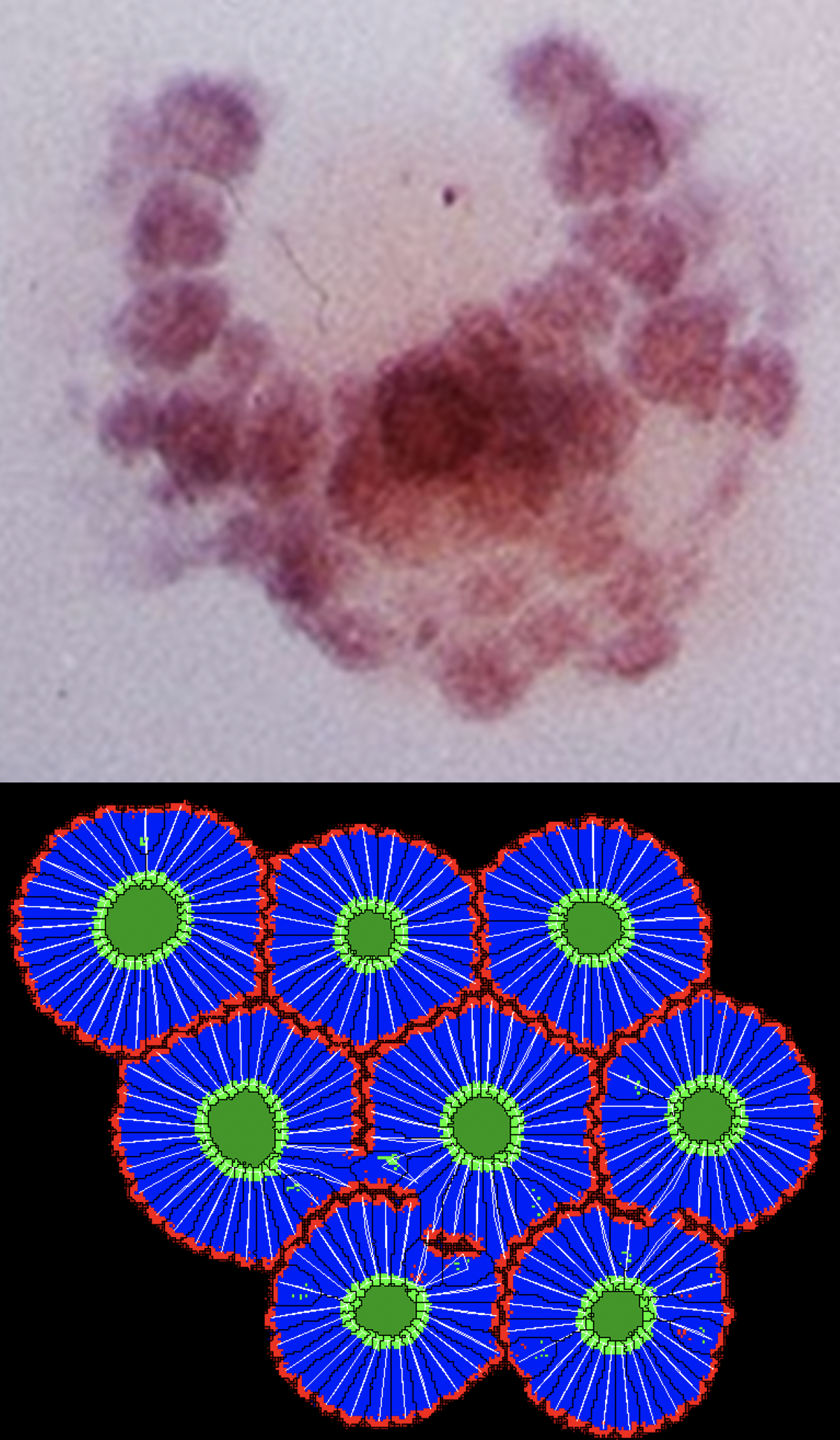
The video below shows side by side simulations of 2 models for somite formation:
Left: Multi-scale simulation of the clock and wavefront mechanisms showing (left) cell adhesion properties, (center) fgf8 wavefront, (right) clock Lfng levels.
Right: The self-organized model of somite formation where cells undergo mesenchymal-to-epitheial transition.
Other lines of research on the topic of developmental biology include computer simulations of:
– Limb-bud formation
– Stomach left-right asymmetry
– Gut morphogenesis
– Drosophila gastrulation
– Drosophila tracheal system development
– Planar Cell Polarity (PCP)
There are open positions for students interested in working on these topics.
Please, contact us!
Check our publications‘ page for a full list of papers.